Things I have done at work, at school, and beyond!
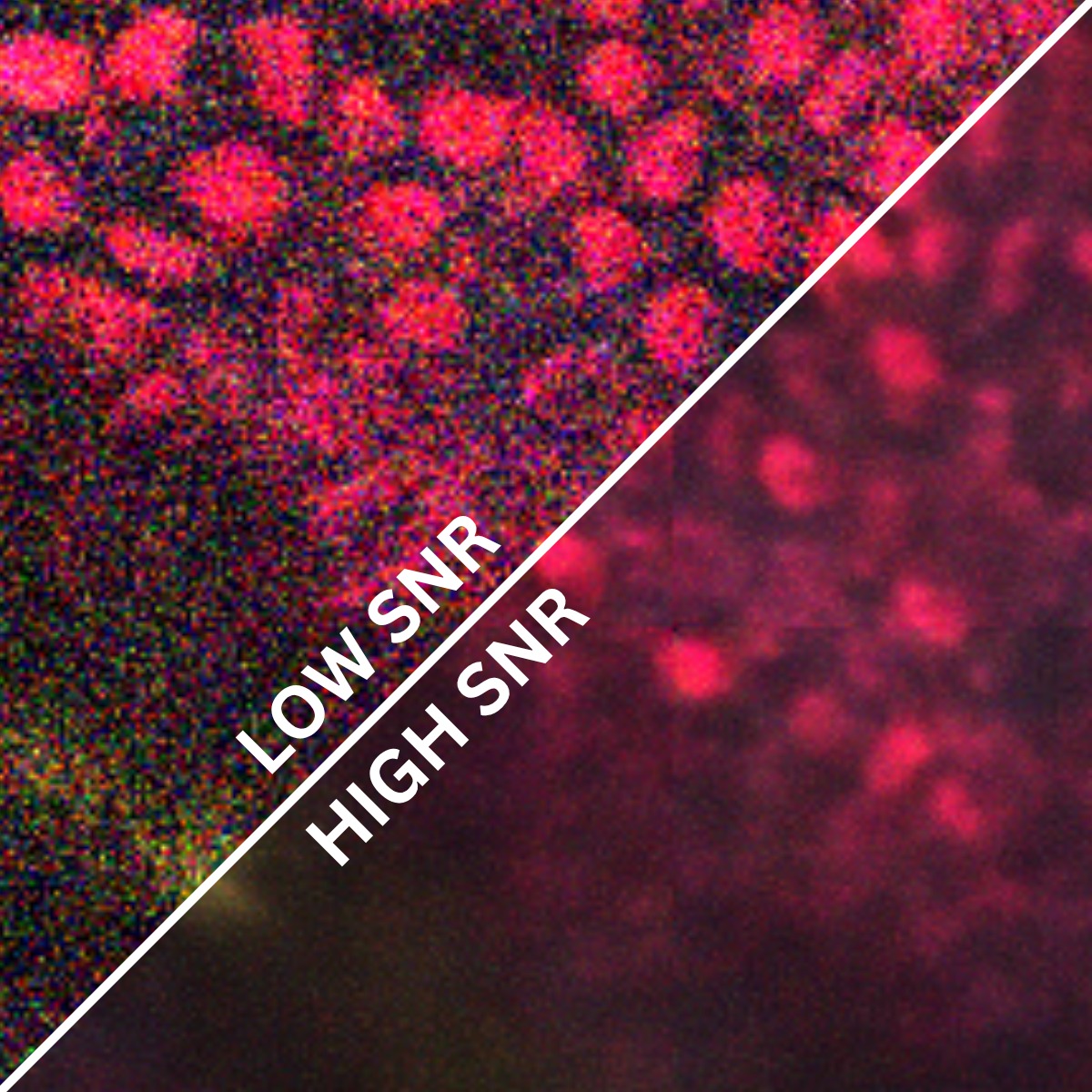
Models & Methods: Dermis & Epidermis • Predictive diagnostics • FLAME microscopy • Content-aware image restoration • Fluorescence lifetime imaging • Computer vision
Languages & Frameworks: Python • Tensorflow • UNet • ONNX • MLFlow • MLOps • OpenCV • WSL • Jupyter • Command-line
Impact:
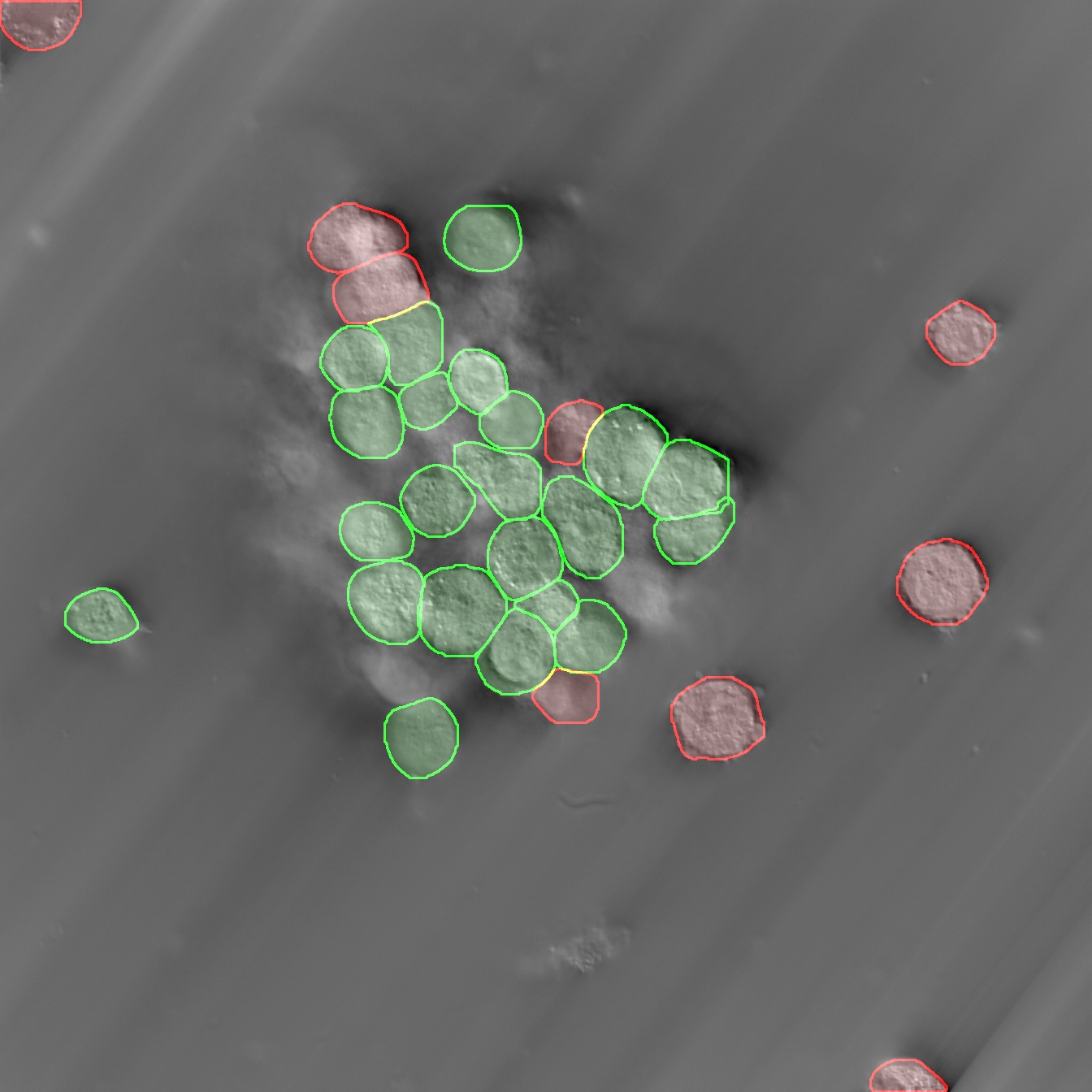
Models & Methods: Mammalian bioreactors • Bioprocess engineering • Instance segmentation • Product development • Digital staining • Cell viability • quantitative phase imaging (QPI) • CHO cells • HEK293 cells • E. coli • S. cerevisiae • UX design
Languages & Frameworks: Python • C++ • Batch • Bash • Tensorflow • PyTorch • Detectron2 • TensorRT • ONNX • Vertex AI • Lightning AI • OpenCV • WSL • Qt
Impact:
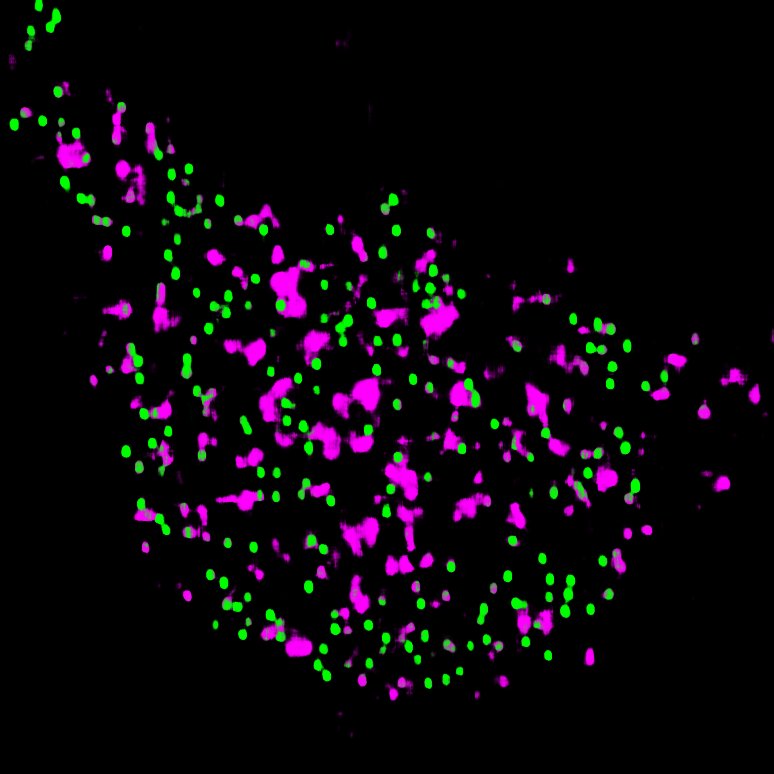
Models & Methods: AI/ML • Computer vision • Phase separation • Aging • Murine primary cortical culture • Murine DRG culture • Airyscan confocal microscopy • Live & fixed immunofluorescence • MTT assay • Extracellular flux analysis • High-performance computing • Plasmid amplification • HEK293 culture • R-Loops
Languages & Frameworks: Python • Tensorflow • Bash • HPC • Slurm • OpenCV • ImageJ / FIJI • Jython
Impact:
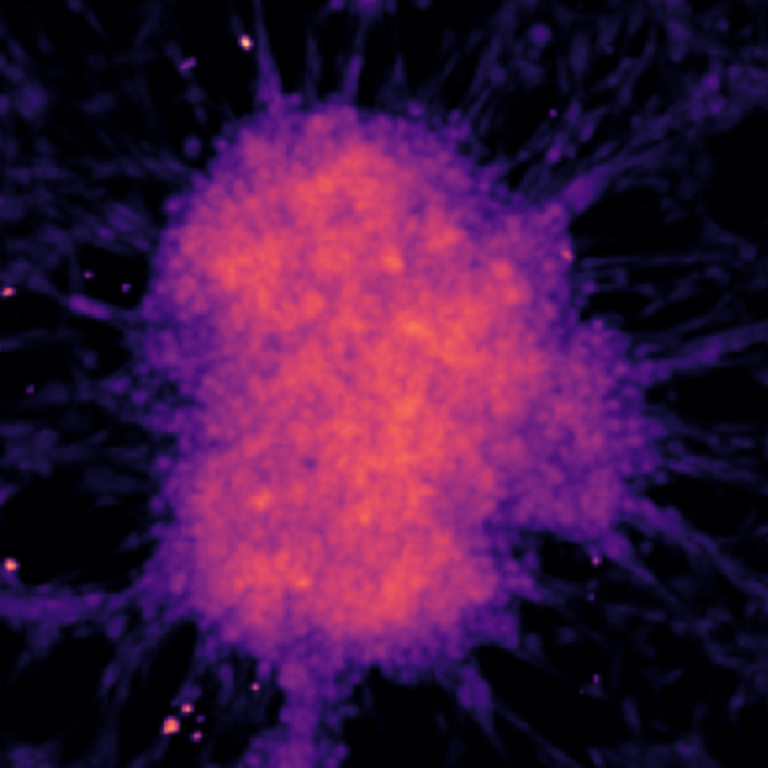
Models & Methods: Glioblastoma multiforme (GBM) • U87 cell line • Western blotting • Crystal violet assay • Genetic code expansion • Redox biology • Protein engineering • Confocal microscopy • Synthetic biology • Python • Jupyter • OpenCV
Languages & Frameworks: Python • Jupyter • OpenCV • Matplotlib
Impact:
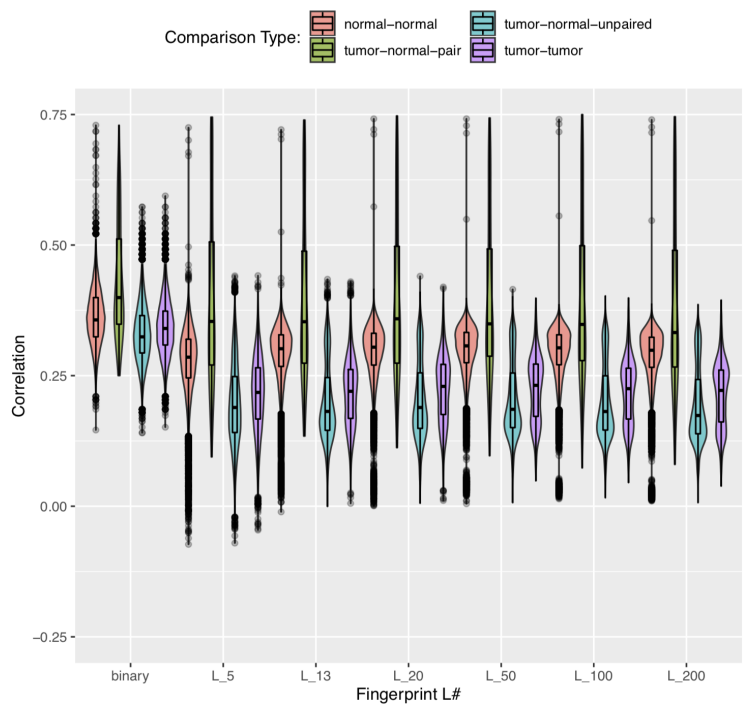
Languages: Python • SQL • R
Frameworks: AWS • GCP • Jupyter • Pandas • RStudio • VCF
Domains: Bioinformatics • Oncology • Cloud Computing • Statistics • Visualization
Destription: Implemented genome fingerprinting and comparison into a custom tissue processing pipeline for patient-personalized CAR-T therapy in the nation's 11th largest health system. Integrated into existing AWS workflow as a quality measure to conclusively determine whether a tumor sample comes from a patient or not.